The GMCF has extensive instrumentation to enable NGS workflows, with a focus on automation and semi-automation. In addition to robotics for nucleic acid extraction, liquid handling, and fully automated NGS library preparation, the GMCF is equipped with ancillary equipment for molecular biology work, listed below.
-
Image

- Applied Biosystems 2 x 96 well MiniAmp Instruments
- Applied Biosystems QuantStudio 5 Real-Time PCR (384-well)
- Applied Biosystems QuantStudio 3 Real-Time PCR (96-well)
- Cole-Parmer Techne PrimePro48 Real Time qPCR
- Qiagen QIAcuity Digital PCR System (4x96-well throughput)
- The QIAcuity Digital PCR System is designed to deliver precise and multiplexed quantification results for mutation detection, copy number variation (CNV), gene expression studies, gene-editing analysis, and many more.
- Thermofisher ProFlex (7x96 instruments); 1xdual-head 384 instrument; 1xtriple-head 32 instrument)
-
Image
 Image
Image
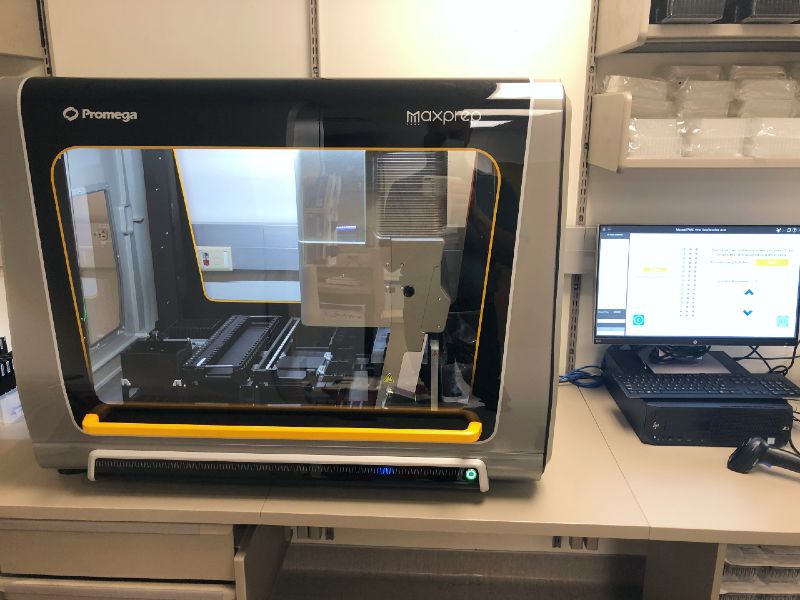
- Promega MaxPrep Liquid Handler
- Qiagen QIAcube Nucleic Acid Extraction Robot
- Qiagen TissueLyser II Bead-beating device
- Promega Maxwell RSC48
- Promega Maxwell RSC16
- Revvity Chemagic 360 Instrument
-
Image
 Image
Image
- Agilent TapeStation4150
- Agilent TapeStation systems are automated electrophoresis solutions for quality control (QC) of DNA and RNA samples.
- Sage Scientific Blue PippinPrep automated DNA size selection. This device allows for gel excision-free size selection of DNA fragments for NGS workflows.
- Invitrogen E-Gel Power snap electrophoresis device and camera
- Invitrogen Qubit Fluorometer
- Promega GloMax Discover Microplate Reader
- Agilent TapeStation4150
-
Image

- Integra VIAFLO96 semi-automated pipettor
- Integra Assist Plus pipetting robot
- Integra Mini 96 pipettor
- OpenTrons OT2 Lab Robot (x3)
- The OT-2 lab robot is an open-source user friendly liquid handling robot to automate tasks such as library prep, ribosomal RNA depletion, poly(A) mRNA enrichment, sample normalization and re-pooling of sequencing libraries.
- Revvity Sciclone® G3 NGSx Workstation
- The Sciclone G3 workstation is a fully automated, high-throughput NGS library preparation system that can process up to 96 samples at a time for DNA and RNA libraries, including standard RNAseq, rRNA depletion, DNAseq, microRNAseq, methyl-seq, hybridization capture, and more. Full automation allows for high reproducibility and rapid turnaround.
- Revvity JANUS
- The Janus is a workstation used for automation of pre-processing of air filter samples for downstream nucleic acid extraction.
- Revvity BioQule
- The BioQule is a fully automated, low-throughput NGS library preparation system that can process up to 8 samples at a time for DNA and RNA libraries. Eventually, the BioQule system is expected to be able to integrate DNA extraction and DNA library preparation into a single automated workflow with minimal hands-on effort required.
-
- 10X CytAssist
- shared with Rush University Histology Core Facility
- Fluent Biosciences
- Cell separation system
- 10X CytAssist
-
The GMCF facility houses several small-to-medium sized sequencers, with big data generation performed externally on Illumina NovaSeq6000 instruments (NovaSeq6000 X coming soon) and on PacBio Sequel II devices. Internal instruments are used for NGS amplicon sequencing and projects with small data needs or those where a short turn-around-time is essential. In addition, most Illumina libraries made at the GMCF facility are initially sequenced on a small sequencing run (‘QC sequencing’) to evaluate library quality and to standardize sequencing depth between samples.
Sequencer
Output (clusters/run)
Maximum Read Length
Notes
Illumina iSeq
4 million
2 x 150 bp
Illumina MiniSeq
8 million
2 x 150 bp
Illumina MiSeq (access only)
Up to 25 million
2 x 300 bp
2 x 250 bp
Standard V2, V3, and Nano flow cells available.
Illumina NovaSeq6000 (access only)
Up to 400 million
2 x 250 bp
SP flow cells available. Excellent alternative to MiSeq sequencing for high throughput samples at a similar cost.
Illumina NovaSeq X Plus
Up to 1 to 3.2 billion
1 x 100 bp
2 x 150 bp
10B and 25B flow cells available.
Oxford Nanopore MinION
Variable
No theoretical upper limit
Dependent on nucleic acid quality. Excellent for long fragment sequencing and de novo assembly.
Oxford Nanopore P2 Solo
Variable
No theoretical upper limit
Dependent on nucleic acid quality. Excellent for long fragment sequencing and de novo assembly.
PacBio Revio
Up to 90 million
No theoretical upper limit
Excellent for full-length 16S and long-read amplicons.
-
- 2D barcode Matrix system, including:
- VisionMate High Speed Barcode Reader
- VisionMate Wireless Reader (x2)
- 2D barcode Matrix system, including:
